Table of Contents (click to expand)
The C-value paradox is the size of the genome (the amount of DNA) doesn’t correlate with how complex an organism is. In other words, the C-value paradox means that a larger genome doesn’t always lead to more complexity.
During a genetics lecture in college, someone asked, “Do humans have the most DNA?”
I thought it was a foolish question. I thought wrong. Our professor chuckled, “Not by a long shot. Actually, a Japanese flower has a genome that is 50 times larger than ours.”
The class, prepared to hear a resounding “yes”, cementing human glory, stared at her in disbelief. How and why would a plant have more DNA than humans, far more evolved and complex mammals? Is there any relationship between the amount of DNA an organism has and its complexity? In other words, does size matter?
The short answer is: yes, it does matter. But also no, it doesn’t.
Recommended Video for you:
What Is The C-value Paradox?
The C-value paradox is that the size of the genome (the amount of DNA) doesn’t correlate with how complex an organism is. In other words, the C-value paradox means that a larger genome doesn’t always lead to more complexity.
One would expect that complexity requires more DNA. The evolution from microscopic life, like bacteria and archaea, to larger and more multiplex life, like humans, requires more information and therefore more DNA.
This is true.
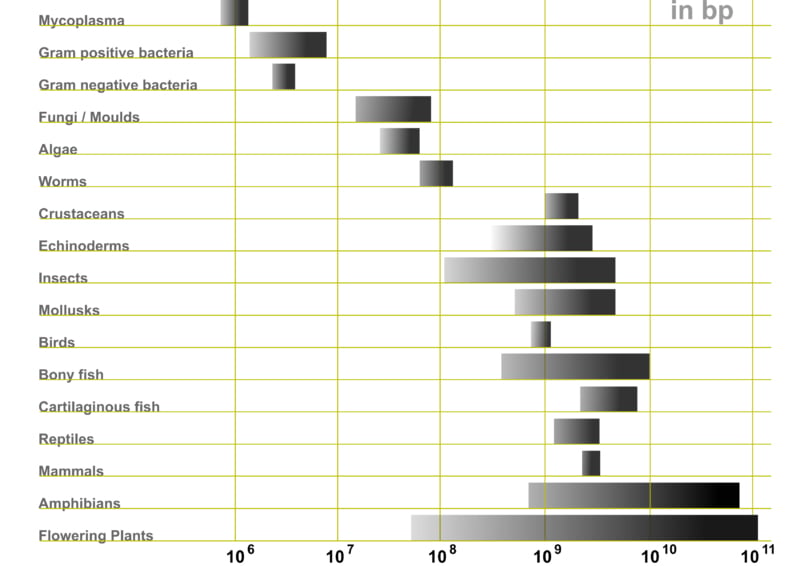
Eukaryotes, organisms with a nucleus to house their genetic material (DNA), do have larger genomes compared to prokaryotes (organisms without a nucleus) such as bacteria. E.coli, every microbiologist’s best friend, has only 4 million base pairs. The fly, Drosophila Melanogaster, on the other hand, has 140 million base pairs. A fly is clearly more complex than a bacteria.
However, this logic doesn’t hold when one compares the genomes of eukaryotes.
Consider humans and the marbled lungfish. The marbled lungfish places second in the ‘largest genome’ contest, with a whopping 130 billion base pairs. Humans, for comparison, only have 4 billion base pairs worth of DNA.
Yet, humans are more complex than marbled lungfish, with our sophisticated physiological systems, modes of communication, movement, etc. Why then does a fish have 40 times as much DNA?
How Was The C-value Paradox Discovered?
In 1971, C.A. Thomas Jr., a geneticist, dubbed this problem the C-value paradox. The C-value is the amount of DNA in a haploid nucleus (measured in picograms), like sperm cells. Regular, autosomal cells have two sets of DNA—one maternal and one paternal. These diploid cells have two chromosomes that work together. In that case, an autosomal human cell has 6 billion base pairs worth of DNA.
Why Does The C-value Paradox Happen?
All DNA isn’t equal.
The entirety of the genome doesn’t hold information crucial for survival.
Genes are the stars of the genome. They are the segments of DNA that code for parts of the cellular machinery, primarily proteins, and RNA. Many scientists assumed that more DNA meant more genes which meant more protein which could do more work to make an animal complex.
But that isn’t the case. There are sections of DNA that don’t contain genes or other parts that help a gene function.
Susume Ohno, a geneticist with an impressive handlebar mustache, called it ‘junk DNA’. In eukaryotes, this supposedly useless DNA takes up a lot of space. Half the human genome is junk, while the marbled lungfish only needs 1/36th of its humongous genome.

What Is Junk DNA?
Junk DNA, as mentioned before, is DNA that doesn’t hold any indispensable information for the organism’s survival and is a result of mutations. Some mutations are monumental, such as whole-genome duplication mutation, where the entire genome gets doubled.
Much of the Junk DNA doesn’t have any direct implication on the complexity of an animal. There is evidence that junk DNA plays some functions in organisms, but to what extent and how much of junk DNA is important for complexity is still unknown.
But, the animals with the largest genomes have a lot of junk DNA that doesn’t seem to contribute to their complexity.
Tl;dr
Eukaryotes have larger genomes than prokaryotes (bacteria), but more complex eukaryotes do not have larger genomes than less complex organisms. This is called the C-value paradox, where the C-value means the amount of DNA in a haploid cell. This is because there is part of the DNA that does not have any informational function, called junk DNA (still debatable). Sometimes, this junk DNA litters an organism’s genome, making it much larger than it needs to be. More complex organisms achieve that level of complexity by regulating their genes better. They do this by alternative splicing, signal transduction and epigenetic mechanisms (amongst many more). This means that their genes have more functions, thus allowing for more complex expression and form.
References (click to expand)
- The size of the genome and the complexity of living beings. metode.org
- Doolittle, W. F., & Brunet, T. D. P. (2017, December). On causal roles and selected effects: our genome is mostly junk. BMC Biology. Springer Science and Business Media LLC.
- The Complexity of Eukaryotic Genomes - The Cell - NCBI. The National Center for Biotechnology Information
- Eddy, S. R. (2012, November). The C-value paradox, junk DNA and ENCODE. Current Biology. Elsevier BV.